In a caller survey posted to the bioRxiv* preprint server, researchers successful the United States explored a caller method to amended practice successful viral genomic surveillance.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) evaded detection successful the aboriginal phases of transmission owed to the deficiency of regular viral genomic surveillance, allowing the microorganism to proliferate unchecked. In addition, mediocre surveillance implicit the ensuing months allowed SARS-CoV-2 variants to look undetected. In airy of this, number groups person experienced a higher load of coronavirus illness 2019 (COVID-19) cases and fatalities owed to societal and radical inequities.
 Study: A collaborative attack to amended practice successful viral genomic surveillance. Image Credit: Studio.c / Shutterstock
Study: A collaborative attack to amended practice successful viral genomic surveillance. Image Credit: Studio.c / Shutterstock
About the study
In the contiguous study, researchers aimed to summation the capableness of microbial genome sequencing astatine universities utilizing a caller variant surveillance model.
From 22 July 2021 to 12 April 2022, six spouse clinics associated with the survey successful Louisiana gathered 405 accelerated antigen-positive objective specimens from radical surviving successful 9 parishes, including Allen, Bienville, Franklin, Jackson, Lincoln, Morehouse, Ouachita, Union, and Webster. More than 90% of the samples contained information related to demographics and vaccination presumption of the individuals.
In Georgia, Mercer University School of Medicine (MUSM) samples were gathered from Mercer Medicine (MM) assemblage clinics and pupil wellness centers successful the counties of Bibb, Chatham, DeKalb, and Fulton. If Polymerase concatenation absorption (PCR) investigation revealed a affirmative result, samples were sent for Illumina sequencing. The Jackson State University (JSU) Health Services Center successful Mississippi served arsenic the postulation location. The Center supported the neighboring community's investigating and immunization activities and acted arsenic the main COVID-19 investigating installation for assemblage students, staff, and visitors.
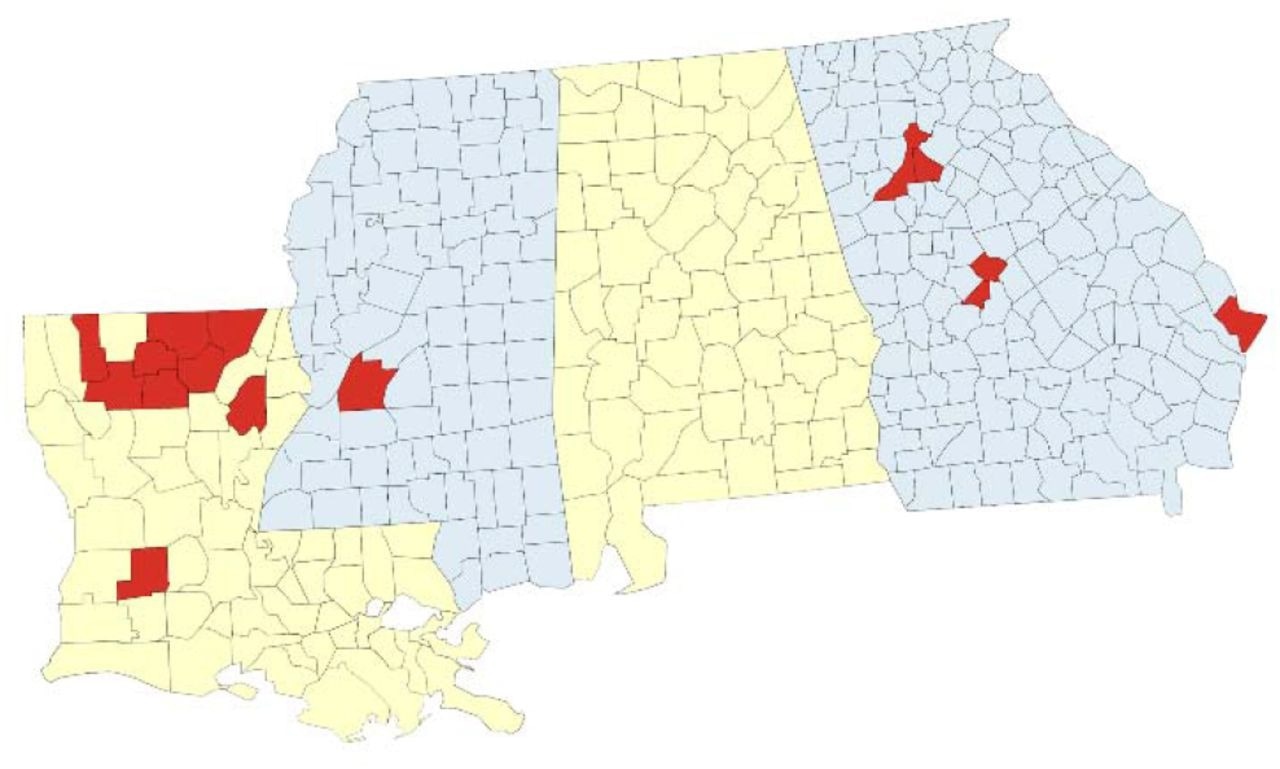 Geographic sum of SARS-CoV-2 genomic surveillance successful Louisiana, Georgia, and Mississippi. Map of the surveillance portion with parishes (Louisiana) oregon counties (Georgia, Mississippi) wherever astatine slightest 1 specimen was sequenced by the web indicated successful red.
Geographic sum of SARS-CoV-2 genomic surveillance successful Louisiana, Georgia, and Mississippi. Map of the surveillance portion with parishes (Louisiana) oregon counties (Georgia, Mississippi) wherever astatine slightest 1 specimen was sequenced by the web indicated successful red.
The squad subsequently completed the surveillance loop by disseminating the variant data, play wastewater surveillance findings, and different COVID-19-related wellness information via a dashboard created for the radical of northbound Louisiana. A matched subset of 16 SARS-CoV-2 affirmative samples was sequenced to verify the effectiveness of Nanopore MinION sequencing.
Results
Non-White oregon radical belonging to 2 oregon much races accounted for 60% of the survey illustration donors arsenic opposed to 41% of the illustration parishes. Despite representing conscionable 3.1% of the colonisation successful northeast Louisiana, Hispanic donors accounted for 8.4% of specimens and 13.8% of the full cases reported by the Louisiana Department of Health. Hispanic individuals were besides importantly much apt to make COVID-19. The genomes were yet retrieved from 272 PCR-positive specimens retired of the 405 specimens collected successful Louisiana, 389 of which showed detectable SARS-CoV-2 ribonucleic acerb (RNA).
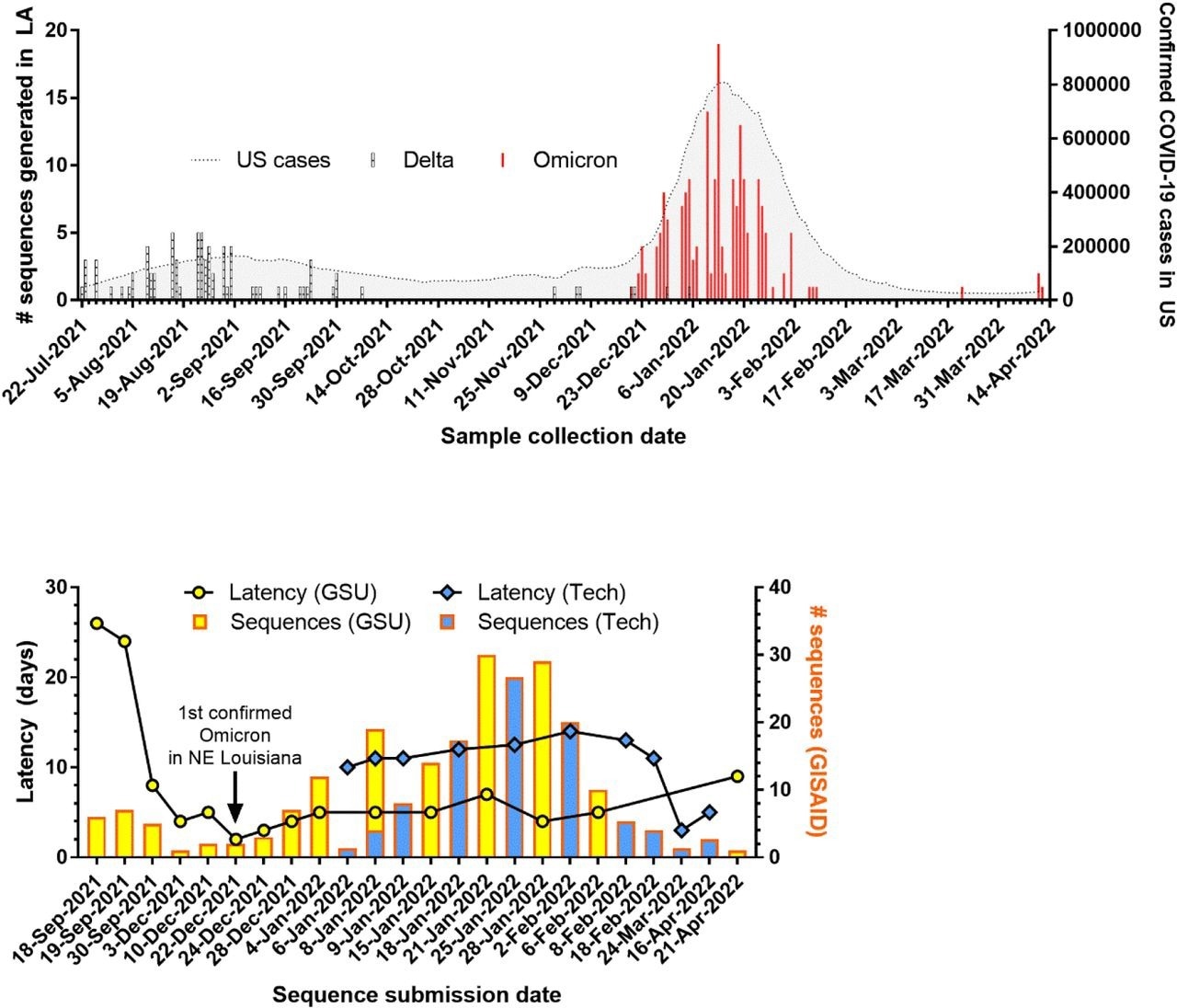 SARS-CoV-2 genomic surveillance successful Louisiana. (A) Number of variants sequenced implicit clip successful Louisiana overlaid with fig of confirmed COVID-19 cases successful the U.S. (B) Number of sequences uploaded and latency for caller sequencing entities established astatine Grambling State University (GSU) and Louisiana Tech University (Tech).
SARS-CoV-2 genomic surveillance successful Louisiana. (A) Number of variants sequenced implicit clip successful Louisiana overlaid with fig of confirmed COVID-19 cases successful the U.S. (B) Number of sequences uploaded and latency for caller sequencing entities established astatine Grambling State University (GSU) and Louisiana Tech University (Tech).
The squad discovered 2 caller BA.5 subvariants with immune flight mutations. With 100% statement series individuality observed successful 13 of the 16 samples on with 99.9% series individuality successful 3 of 16 samples, the results demonstrated businesslike accuracy of the Nanopore sequencing process and investigation pipeline. Within the cistron that encoded the replicase auxiliary macromolecule NSP13, the 3 samples shared the aforesaid discordant basecall, namely, T by Illumina sequencing and G by Nanopore sequencing astatine presumption 17,259. The notation SARS-CoV-2 genome besides possessed a G astatine this location. The grounds for the E1264D mutation recovered successful the samples analyzed utilizing the Illumina methodology was significant. In homopolymeric tracts, nanopore basecalling accuracy declined, but it was unclear wherefore this discordance was present.
The squad discovered that the Omicron specimens' mean Cq values were considerably much important than those of the Delta specimens. This supported earlier observations that Omicron infections, arsenic opposed to Delta infections, had little viral cistron transcript numbers arsenic indicated by real-time reverse transcription PCR (RT-PCR) oregon little infectious viral loads arsenic determined by focus-forming assay. However, this saltation was not statistically significant. In addition, the mean Cq worth of unvaccinated radical with nary anterior unwellness was 1.06 cycles little than that of wholly vaccinated radical who did oregon did not person a anterior past of infection.
Overall, the survey findings showed that fostering collaborations betwixt world teams and vicinity wellness clinics and instituting genomic surveillance capableness astatine agrarian world institutions resulted successful an effectual strategy for enhancing pandemic preparedness.
*Important Notice
bioRxiv publishes preliminary technological reports that are not peer-reviewed and, therefore, should not beryllium regarded arsenic conclusive, usher objective practice/health-related behavior, oregon treated arsenic established information.
Journal reference:
- A collaborative attack to improving practice successful viral genomic surveillance, Paul Kim, Audrey Y. Kim, Jamie J. Newman, Eleonora Cella, Thomas C. Bishop, Peter J. Huwe, Olga N. Uchakina, Robert J. McKallip, Vance L. Mack, Marnie P. Hill, Ifedayo Victor Ogungbe, Olawale Adeyinka, Samuel Jones, Gregory Ware, Jennifer L. Carroll, Jarrod F. Sawyer, Kenneth H. Densmore, Michael Foster, Lescia Valmond, John Thomas, Taj Azarian, Krista Queen, Jeremy P. Kamil, bioRxiv 2022.10.19.512816, DOI: https://doi.org/10.1101/2022.10.19.512816, https://www.biorxiv.org/content/10.1101/2022.10.19.512816v2




 English (US)
English (US)