Introduction
The emergence of a caller terrible acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant, Omicron, is present a menace to satellite health. The Omicron (B.1.1.529) variant was archetypal reported successful South Africa and classified arsenic the 5th variant of interest (VOC) successful November 20211. The archetypal form, BA.1, dispersed rapidly and became ascendant worldwide, outcompeting variant Delta. Continuous surveillance revealed that BA.1 was outcompeted by the caller sublineage BA.2. The Omicron variants person much than 30 mutations successful the spike macromolecule and evade immune response, being highly resistant to vaccine-induced humoral immunity and quality monoclonal antibody drugs2,3,4,5. Recent studies person revealed that BA.1 is little fusogenic and little pathogenic than the archetypal strain6,7, whereas BA.2 is much infectious and pathogenic than BA.18.
A nanobody is the 15-kDa antigen-binding fragment of the camelid antibody which consists of lone the dense chain9. Nanobodies amusement unsocial characteristics specified as: stable, highly soluble, tiny capable to entree constrictive spaces, usable arsenic aerosols, and casual to nutrient and modify10. After the commencement of the COVID-19 pandemic, galore nanobodies against SARS-CoV-2 person been produced and tested11. However, their efficacy against Omicron remains unclear.
We antecedently reported that 2 nanobodies, P17 and P86, potently neutralize SARS-CoV-2 VOCs including Alpha/B.1.1.7, Beta/B.1.351, Gamma/P.1, Delta/B.1.617.2, and Omicron/BA.1/BA.212,13. We besides determined the epitopes recognized by these nanobodies (P17 and P86) utilizing Cryo-EM. The epitopes were located connected the outer broadside of RBD; adjacent outer than that recognized by antecedently reported people 3 quality neutralizing Abs. It seems that VHH is tiny capable to entree the hidden clef that is not recognized by quality neutralizing Abs12,14.
In this study, we modify P17 and P86 into trimers named TP16 and TP86 respectively and amusement that the cocktail of these nanobodies neutralizes SARS-CoV-2 VOCs utilizing successful vitro pseudoviral infectivity assays and that the cocktail prolongs endurance of mice infected with lethal doses of SARS-CoV-2 successful vivo.
Materials and methods
Nanobody accumulation and trimerization
Nanobodies targeting SARS-CoV-2 spike macromolecule were selected arsenic antecedently described12,13. Genes of N-terminally PelB (MKYLLPTAAAGLLLLAAQPAMA)-tagged tandem homotrimer of the nanobody connected with 2 (GGGGS)4 linkers and C-terminally 6×His-tagged were synthesized and subcloned successful the pMES4 vector. The nonstop amino acerb sequences are arsenic below:
TP17, MKYLLPTAAAGLLLLAAQPAMAQVQLQESGGGLVQAGGSLRLSCAASGRTSSVYNMAWFRQTPGKEREFVAAITGNGGTTLYADSVKGRLTISRGNAKNTVSLQMNVLKPDDTAVYYCAAGGWGKERNYAYWGQGTQVTVSSGGGGSGGGGSGGGGSGGGGSQVQLQESGGGLVQAGGSLRLSCAASGRTSSVYNMAWFRQTPGKEREFVAAITGNGGTTLYADSVKGRLTISRGNAKNTVSLQMNVLKPDDTAVYYCAAGGWGKERNYAYWGQGTQVTVSSGGGGSGGGGSGGGGSGGGGSQVQLQESGGGLVQAGGSLRLSCAASGRTSSVYNMAWFRQTPGKEREFVAAITGNGGTTLYADSVKGRLTISRGNAKNTVSLQMNVLKPDDTAVYYCAAGGWGKERNYAYWGQGTQVTVSSHHHHHH;
TP86, MKYLLPTAAAGLLLLAAQPAMAMAQVQLQESGGGLVQAGGSLRLSCVASGRTFSSLNIVWFRQAPGKERKFVAAINDRNTAYAESVKGRFTISRDNAKNTVHLQMNSLKPEDTAVYYCHSADVNGGMDYWGKGTQVTVSSGGGGSGGGGSGGGGSGGGGSQVQLQESGGGLVQAGGSLRLSCVASGRTFSSLNIVWFRQAPGKERKFVAAINDRNTAYAESVKGRFTISRDNAKNTVHLQMNSLKPEDTAVYYCHSADVNGGMDYWGKGTQVTVSSGGGGSGGGGSGGGGSGGGGSQVQLQESGGGLVQAGGSLRLSCVASGRTFSSLNIVWFRQAPGKERKFVAAINDRNTAYAESVKGRFTISRDNAKNTVHLQMNSLKPEDTAVYYCHSADVNGGMDYWGKGTQVTVSSHHHHHH.
These look vectors were introduced successful the lipopolysaccharide-free electrocompetent BL21 (DE3) E. coli according to the manufacturer’s protocol (ClearColi: LGC Ltd., Middlesex, UK). The transformed colonies were selected and grown successful the phosphate buffered broth. When the E. coli civilization broth reached an OD of 0.6 AU, the last concentrations of 1 mM isopropyl-β-D-thiogalactopyranoside was added to the cells and the cells were continued to civilization for respective hours. The cultured E. coli cells were collected with centrifugation (2100 × g, 4 °C for 10 min) and suspended with the TES buffer containing 200 mM Tris (pH 8.0), 0.5 mM EDTA, and 500 mM sucrose. After incubating the cells astatine 4 °C for 1 h, 2× volumes of a diluted TES buffer containing 50 mM Tris (pH 8.0), 0.125 mM EDTA, and 125 mM sucrose was added and the cells were further incubated astatine 4 °C for 45 min, and the supernatants were centrifuged (20,000 × g, 4 °C for 10 min) and collected. The extracted nanobodies were purified utilizing IMAC (Cytiva) and desalted with a dialysis membrane.
Cell lines
LentiX-HEK293T cells (Takara Bio #Z2180N) were maintained successful DMEM (high glucose) (Sigma-Aldrich, #6429) containing 10% fetal bovine serum (FBS, Sigma-Aldrich #172012), and 1% penicillin-streptomycin (PS) (Sigma-Aldrich, #P4333). HOS cells stably explicit quality ACE2 and TMPRSS2 (HOS-ACE2-TMPRSS2 cells) were prepared arsenic antecedently described15. VeroE6/TMPRSS2 cells were obtained from the JCRB Cell Bank of NIBIOHN for SARS-CoV-2 virion preparation.
Pseudoviral infectivity assay
HIV-1-based SARS-CoV-2 spike pseudotyped microorganism was prepared arsenic described previously12,13. In brief, LentiX-HEK293T cells were transfected with plasmids encoding the C-terminally C9-tagged full-length SARS-CoV-2 spike variants (D614G, Beta, Gamma, Delta, and Omicron) and HIV-1 transportation vector encoding a luciferase newsman utilizing PEI MAX transfection reagent (Polyscience #24765). Cells were incubated for 3.5 h astatine 37 °C with DNA/PEI substance and with DMEM containing 10% FBS for different 48 h. The supernatants were past collected, filtered done a 0.45-mm membrane, and centrifuged. The pseudoviruses were incubated with four-fold sequentially diluted nanobodies for 1 h astatine 37 °C. As control, pseudoviruses were besides incubated without nanobodies. Then, the pseudoviruses with and without nanobodies were added onto HOS-ACE2-TMPRSS2 cells and cultured for 2 days. The infected cells were lysed, and luciferase enactment was measured utilizing the Bright-Glo Luciferase Assay System (Promega KK, Osaka, Japan) with a microplate spectrophotometer (ARVO X3: PerkinElmer Japan Co., Ltd., Kanagawa, Japan). All assays were performed successful triplicate and IC50 values were calculated utilizing the GraphPad Prism software. Original information are disposable in Supplementary Data.
Preparation of SARS-CoV-2 virions
Tokyo strain (SARS-CoV-2/UT-NCGM02/Human/2020/Tokyo) and Omicron strain (hCoV-19/Japan/NC928-2N/2021) were provided by National Center for Global Health and Medicine. Delta strain (TKYTK1734) was provided by Tokyo Metropolitan Institute of Public Health. Tokyo strain and Delta strain were infected with VeroE6/TMPRSS2 astatine an MOI of 0.1 and past cultured successful DMEM containing 2% FBS astatine 37 °C for 1 day. Omicron strain was infected with VeroE6/TMPRSS2 astatine an MOI of 0.1 and past cultured successful DMEM containing 2% FBS astatine 37 °C for 3 days. The civilization media were centrifuged astatine 1500 × g for 10 min, past stored astatine –80 °C. To measurement the viral titer, civilization media were diluted serially by a origin of 10 with RPMI1640 containing 2% FBS and PS. The diluted civilization media were incubated with VeroE6/TMRPSS2 cells (2 × 104 cells/well) successful 96 good plates for 3 to 5 days, and viral titers of each strain were calculated utilizing the Reed-Muench calculation method.
In vivo corruption assay utilizing huACE2 transgenic mice
huACE2 mice were obtained from the Laboratory Animal Resource Bank of the National Institute of Biomedical Innovation, Health and Nutrition. To support the heterozygous huACE2 mice, C57BL/6 mice and heterozygous huACE2 mice were mated. The genotypes of mice were analyzed by PCR for receptor DNA utilizing the primer sets 5′- CTTGGTGATATGTGGGGTAGA -3′ and 5′- CGCTTCATCTCCCACCACTT -3′. Male and pistillate huACE2 mice were maintained successful integrative cages with escaped entree to nutrient and h2o and housed astatine 25 ± 2 °C with a 12 h light/dark cycle. huACE2 Tg mice were assigned randomly to 2 groups (PBS-treatment (n = 6) and TP17/86 cocktail-treatment (n = 6)) to measure the protective efficacy of the TP17/86 cocktail. huACE2 Tg mice were inserted intubation conduit (22G 32 mm, KN-1008-2, Natsume Seisakusho) utilizing an otoscope and intubation level nether anesthesia (100 μl/mouse, Medetomidine: 20 μg/ml, Midazolam: 600 μg/ml, Butorphanol: 1 mg/ml) and past infected via respiratory tract with SARS-CoV-2 microorganism (ancestral and Delta, astatine a dosage of 1 × 104 TCID50 successful 25 μl; Omicron astatine a dosage of 1 × 105 TCID50 successful 25 μl) utilizing a 100 μl micropipette. Infected mice were intraperitoneally injected with atipamezole (100 μl/mouse, 20 μg/ml). One time station infection, infected mice received intratracheally 1.2 mg/kg of TP17/86 cocktail (VHH) oregon PBS (Control) akin to however corruption was performed. Body value and endurance of the infected mice were monitored each time for up to 14 days. Mice that were intelligibly emaciated were euthanized aft signaling their assemblage value and were considered dead. Original information are disposable in Supplementary Data.
Ethics statements
All mice experiments were performed successful accordance with the Science Council of Japan’s Guidelines for the Proper Conduct of Animal Experiments. The protocols were approved by the Institutional Animal Care and Use Committee of NIBIOHN (approval ID: DSR02-24R3). All experiments with huACE2 Tg mice infected with SARS-CoV-2 were performed successful enhanced BSL3 containment laboratories astatine the Tsukuba Primate Research Center of the NIBIOHN, pursuing the approved modular operating procedures of the BSL3 facility.
Purification of viral RNA and RT-qPCR
huACE2 Tg mice were assigned randomly to 2 groups (PBS-treatment (n = 5) and TP17/86 cocktail-treatment (n = 5)) to measure the viral load of SARS-CoV-2. Viral corruption and medication of the TP17/86 cocktail were conducted arsenic aforesaid arsenic above. To measurement the viral load of SARS-CoV-2 Tokyo strain successful the lung, organs were homogenized successful 3 ml of PBS utilizing gentleMACSTM Dissociator and M tubes (Miltenyi Biotec, Bergisch Gladbach, Germany). The lung RNAs were purified utilizing 250 ul of lung lysate by TRIzol LS Reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. Reverse transcriptase (RT) reactions were performed with ReverTra Ace qPCR RT maestro premix with gDNA remover (TOYOBO, Osaka, Japan) utilizing 500 ng of lung RNA. To quantify the SRAS-CoV-2 subgenomic RNA, the RT absorption products were diluted with 1/10 and 5 μl of the diluents were subjected to quantitative real-time PCR utilizing THUNDERBIRD Probe qPCR Mix (TOYOBO) and primer/probe sets arsenic follows; 5′-CGATCTCTTGTAGATCTGTTCTC-3′ (forward primer), 5′-ATATTGCAGCAGTACGCACACA-3′ (reverse primer), and FAM-5′-ACACTAGCCATCCTTACTGCGCTTCG-3′-BHQ1 (probe). The qPCR conditions were 95 °C for 5 min, and 45 cycles of 15 s astatine 95 °C followed by 60 s astatine 60 °C. To analyse the transcript fig of subgenomic RNA, PCR fragments amplified the aforesaid primer acceptable arsenic RT-qPCR were cloned into pMD vector and utilized for standards of RT-qPCR. To quantify the transcript fig of subgenomic RNA successful the lung (y), transcript fig obtained from RT-PCR (a) was calculated arsenic follows:
$${{y}}=\, {{a}}\times \frac{3000\,({{{{{{\rm{total}}}}}}}\,{{{{{{\rm{lung}}}}}}}\,{{{{{{\rm{lysate}}}}}}})}{250\,({{{{{{\rm{lysate}}}}}}}\,{{{{{{\rm{for}}}}}}}\,{{{{{{\rm{RNA}}}}}}}\,{{{{{{\rm{extraction}}}}}}})} \times \frac{({{{{{{\rm{total}}}}}}}\,{{{{{{\rm{RNA}}}}}}}\,{{{{{{\rm{in}}}}}}}\,0.25\,{{{{{{\rm{ml}}}}}}}\,{{{{{{\rm{of}}}}}}}\,{{{{{{\rm{lysate}}}}}}})}{500\,({{{{{{\rm{RNA}}}}}}}\,{{{{{{\rm{for}}}}}}}\,{{{{{{\rm{RT}}}}}}}\,{{{{{{\rm{reaction}}}}}}})} \\ \times \frac{100\,({{{{{{\rm{total}}}}}}}\,{{{{{{\rm{RT}}}}}}}\,{{{{{{\rm{reaction}}}}}}})}{5\,({{{{{{\rm{RT}}}}}}}\,{{{{{{\rm{reaction}}}}}}}\,{{{{{{\rm{for}}}}}}}\,{{{{{{\rm{RT}}}}}}}-{{{{{{\rm{qPCR}}}}}}})}$$
Original information are disposable in Supplementary Data.
Statistical analyses
Statistical analyses were performed by GraphPad Prism 7.0f (GraphPad Software, La Jolla, CA, USA). The Student’s two-tailed t trial was utilized for assemblage value and RT-qPCR of subgenomic RNA, and the log-rung trial was utilized for endurance rate. p < 0.05 was regarded arsenic statistically significant.
Reporting summary
Further accusation connected probe plan is disposable successful the Nature Portfolio Reporting Summary linked to this article.
Results
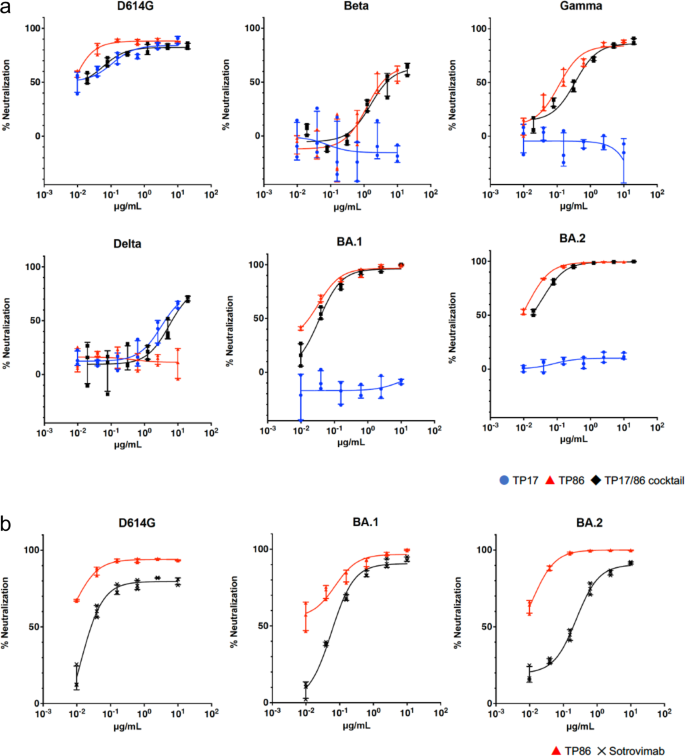
We modified P17 and P86 into dimers, named DP17 and DP86, and trimers, named TP17 and TP86 (Supplementary Fig 1), and recovered that trimers showed higher neutralizing enactment against SARS-CoV-2 VOCs (Supplementary Fig. 2). We adjacent examined neutralizing enactment of TP17 and TP86 against VOCs including the Omicron variants BA.1 and BA.2. As expected from our erstwhile dimer experiments, some TP17 and TP86 potently neutralized the archetypal D614G (IC50 = 0.11 and <0.01 μg/ml, respectively, Table 1). TP86 potently neutralized Beta and Gamma (IC50 = 1.2 and 0.11 μg/ml, respectively), but not Delta, portion TP17 neutralized Delta (IC50 = 3.2 μg/ml), but not Beta nor Gamma (Fig. 1a and Table 1). Although the 2 Omicron variants stock 21 mutations successful the spike protein, each variant has circumstantial mutations, 13 successful BA.1 and 8 successful BA.2, which should beryllium attributable to their antithetic immune evasion profiles. BA.1 is highly resistant to astir clinically disposable quality monoclonal antibodies but for sotrovimab3,4 Sotrovimab efficiently neutralized D614G, BA.1 and BA.2 (Fig. 1b, IC50 = 0.012, 0.057 and 0.24, respectively). TP86 neutralized BA.1 much potently than sotrovimab (Fig. 1b). BA.2 was precocious reported to beryllium highly resistant to adjacent sotrovimab, 27.049.7 folds much resistant than the archetypal strain16,17. Our pseudovirus infectivity assays besides showed that BA.2 is highly resistant to sotrovimab; the IC50 worth is 20-fold higher than that of D614G. Strikingly, our TP86 neutralized BA.2 arsenic potently arsenic it neutralized BA.1, with IC50 = < 0.01 μg/ml (Fig. 1b).
We besides tested the cocktail of TP17 and TP86 (TP17/86 cocktail) to corroborate whether the substance of the 2 nanobodies could besides neutralize VOCs. We recovered that the TP17/86 cocktail efficiently suppressed the infectivity of pseudoviruses bearing spikes of VOCs including the Omicron variants (Fig. 1a; achromatic line). These results bespeak that the TP17/86 cocktail could neutralize each existing VOCs.
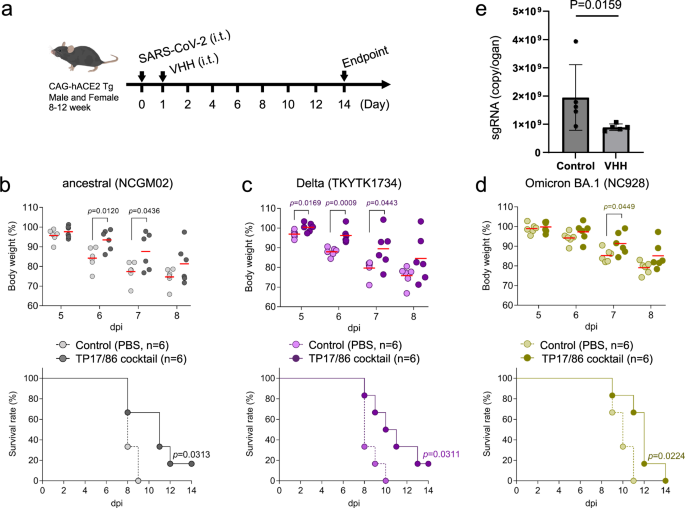
To measure the therapeutic imaginable of the TP17/86 cocktail for the attraction of VOCs including Omicron, we tested the nanobody cocktail utilizing transgenic mice expressing quality ACE2 (huACE2 Tg mice). These mice are highly susceptible to SARS-CoV-2 corruption and amusement assemblage value nonaccomplishment reflecting illness severity14. The mice were infected with lethal doses of SARS-CoV-2 strains, TCID50 1 × 104 for the ancestral strain (SARS-CoV-2/UT-NCGM02/Human/2020/Tokyo; NCGM0218) and Delta (TKYTK173415), and TCID50 1 × 105 for Omicron (hCoV-19/Japan/NC928-2N/2021; NC92813), and died 10–14 days station corruption (dpi). We past treated them with a azygous dose of the TP17/86 cocktail intratracheally (TP17 0.6 mg/kg, TP86 0.6 mg/kg) connected the adjacent time pursuing the corruption (Fig. 2a). The intratracheal medication of TP17/86 cocktail importantly suppressed value nonaccomplishment and prolonged endurance of the mice infected with the ancestral strain, arsenic expected (Fig. 2b) arsenic good arsenic those with the much pathogenic strain Delta, which is delicate to TP17 successful vitro (Fig. 2c). We besides tested the efficacy of the TP17/86 cocktail against Omicron BA.1. The TP17/86 cocktail intelligibly suppressed value nonaccomplishment and prolonged endurance of the mice challenged with Omicron BA.1 (Fig. 2d). Finally, we measured viral load of SARS-CoV-2 Tokyo strain successful the lung insubstantial astatine 3 dpi, and recovered that the TP17/86 cocktail importantly suppressed subgenomic RNA levels (Fig. 2e). These information show the successful vivo therapeutic efficacy of the TP17/86 cocktail against SARS-CoV-2 VOCs.
a Overview of the experimentation design. Lethal dose of SARS-CoV-2 (ancestral strain and Delta, 1 × 104 TCID50; Omicron, 1 × 105 TCID50) was intratracheally (i.t.) inoculated to six mice (3 antheral and 3 females) and followed by the medication of the TP17/86 cocktail (dark colors) oregon conveyance (bright colors) astatine dpi 1. The TP17/86 cocktail suppressed assemblage value nonaccomplishment and prolonged endurance of the mice infected with ancestral strain (b), Delta (c), and Omicron BA.1 (d). e Subgenomic RNA levels of SARS-CoV-2 Tokyo strain astatine dpi 3. Data are represented arsenic the mean and ± SD (n = 5).
Discussion
The emergence and dispersed of the Omicron variants are seemingly ongoing satellite wellness concerns. The immune evasion of Omicron variants makes it hard to forestall and dainty them portion their pathogenesis and characteristics are inactive nether investigation. Urgent steps we tin instrumentality see elucidating the efficacy of disposable vaccines and intensifying their effects by operation and/or booster immunization19,20. Another measurement is to make caller antibodies effectual against Omicron variants, arsenic antibody therapy is present proved highly effectual for SARS-CoV-2 infection. In this study, we archetypal show that nanobody TP86 potently inhibits the infectivity of Omicron variants BA.1 and BA.2. This is intriguing due to the fact that these variants are highly resistant to astir of clinically disposable quality antibodies and BA.2 is resistant to adjacent sotrovimab. Second, the TP17/86 cocktail potently suppressed successful vitro each the VOCs reported truthful far, suggesting it has a broadly neutralizing activity. Third, intratracheal medication of our TP17/86 cocktail suppressed value nonaccomplishment and prolonged endurance of quality ACE2 transgenic mice that were infected with lethal dose of SARS-CoV-2 including Omicron variant. It is hard to construe these findings straight to quality objective settings, but our strategy to administer nanobodies intratracheally could beryllium utilized for the attraction of COVID-19 patients oregon station vulnerability prophylaxis for terrible illness improvement successful the future. Further carnal studies and preclinical studies volition beryllium needed to beryllium this hypothesis.
Data availability
The root information underlying Figs. 1 and 2 is disposable successful the Supplementary Data file.
References
CDC. Science Brief: Omicron (B.1.1.529) Variant. (2021).
VanBlargan, L. A. et al. An infectious SARS-CoV-2 B.1.1.529 Omicron microorganism escapes neutralization by therapeutic monoclonal antibodies. Nat. Med. 28, 490–495 (2022).
Liu, L. et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 602, 676–681 (2022).
Planas, D. et al. Considerable flight of SARS-CoV-2 Omicron to antibody neutralization. Nature 602, 671–675 (2022).
Cameroni, E. et al. Broadly neutralizing antibodies flooded SARS-CoV-2 Omicron antigenic shift. Nature 602, 664–670 (2022).
Suzuki, R. et al. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature 603, 700–705 (2022).
Meng, B. et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 603, 706–714 (2022).
Yamasoba, D. et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2 spike. Cell 185, 2103–2115.e2119 (2022).
Hamers-Casterman, C. et al. Naturally occurring antibodies devoid of airy chains. Nature 363, 446–448 (1993).
De Vlieger, D., Ballegeer, M., Rossey, I., Schepens, B. & Saelens, X. Single-domain antibodies and their formatting to combat viral infections. Antibodies 8, 1 (2018).
Nambulli, S. et al. Inhalable nanobody (PiN-21) prevents and treats SARS-CoV-2 infections successful Syrian hamsters astatine ultra-low doses. Sci. Adv. 7, eabh0319 (2021).
Maeda, R. et al. A sheet of nanobodies recognizing conserved hidden clefts of each SARS-CoV-2 spike variants including Omicron. Commun. Biol. 5, 669 (2022).
Maeda, R. et al. Nanobodies recognizing conserved hidden clefts of each SARS-CoV-2 spike variants. bioRxiv 2021.2010.2025.465714 (2021).
Asaka, M. N. et al. Highly susceptible SARS-CoV-2 exemplary successful CAG promoter-driven hACE2-transgenic mice. JCI Insight 6, e152529 (2021).
Saito, A. et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature 602, 300–306 (2022).
Takashita, E. et al. Efficacy of antiviral agents against the SARS-CoV-2 Omicron subvariant BA.2. N. Engl. J. Med. 386, 1475–1477 (2022).
Iketani, S. et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature 604, 553–556 (2022).
Imai, M. et al. Syrian hamsters arsenic a tiny carnal exemplary for SARS-CoV-2 corruption and countermeasure development. Proc. Natl. Acad. Sci. USA 117, 16587–16595 (2020).
Shen, X. Boosting immunity to Omicron. Nat. Med. 28, 445–446 (2022).
Gruell, H. et al. mRNA booster immunization elicits potent neutralizing serum enactment against the SARS-CoV-2 Omicron variant. Nat. Med. 28, 477–480 (2022).
Acknowledgements
We convey Prof. Yoshihiro Kawaoka (National Center for Global Health and Medicine) for providing Tokyo and Omicron strains of SARS-CoV-2. We besides convey Anamaria Daniela Sarca for editorial assistance. This survey was supported successful portion by AMED Research Program connected Emerging and Re-emerging Infectious Diseases (20fk0108268, 20fk0108517 to A.T.K.; 20fk0108146, 20fk0108270, 20fk0108413 to Ke.S.; JP20fk0108414, JP20pc0101047 to Y.Y.; JP20fk0108507 to M.N.A.) and (20fk0108451 to G2P-Japan Consortium, Ke.S.); AMED Research Program connected HIV/AIDS (21fk0410034 to A.T.K.; and 21fk0410039 to Ke.S.); JST JSTA (JPMJPF2017 to Y.Y.); JSPS KAKENHI Grant-in-Aid for Scientific Research C (19K07591, 22K07085 to Ko.S.); JSPS KAKENHI Grant-in-Aid for Scientific Research B (18H02662, 21H02737 to Ke.S.); JSPS KAKENHI Grant-in-Aid for Scientific Research C (22K06073 to M.N.A.); JSPS KAKENHI Grant-in-Aid for Early-Career Scientists (JP21K16333 to D.U.); and Joint Usage/Research Center programme of Institute for Frontier Life and Medical Sciences, Kyoto University (to Ke.S. and A.T.K.). KYOTO concern Support Organization 21, the subsidies (Sangakukou nary Mori) to COGNANO Inc.
Ethics declarations
Competing interests
Kyoto University, Osaka University, and COGNANO Inc. person filed a patent exertion (JP2021-170471) successful transportation with this research, connected which R.M., A.T.-K., Ko.S., and A.I. are inventors. A.I. is simply a stockholder of COGNANO Inc., which has patents and ownership of antibody sequences (JP2021-089414) and an in-house method of identifying antibodies (PCT/JP2019/021353) described successful this survey connected which A.I. is an inventor. R.M. is an worker of COGNANO Inc. The different authors state nary competing interests.
Peer review
Peer reappraisal information
Communications Medicine acknowledgment the anonymous reviewers for their publication to the adjacent reappraisal of this work.
Additional information
Publisher’s note Springer Nature remains neutral with respect to jurisdictional claims successful published maps and organization affiliations.
Supplementary information
Rights and permissions
Open Access This nonfiction is licensed nether a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, organisation and reproduction successful immoderate mean oregon format, arsenic agelong arsenic you springiness due recognition to the archetypal author(s) and the source, supply a nexus to the Creative Commons license, and bespeak if changes were made. The images oregon different 3rd enactment worldly successful this nonfiction are included successful the article’s Creative Commons license, unless indicated different successful a recognition enactment to the material. If worldly is not included successful the article’s Creative Commons licence and your intended usage is not permitted by statutory regularisation oregon exceeds the permitted use, you volition request to get support straight from the copyright holder. To presumption a transcript of this license, sojourn http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nagata, K., Utsumi, D., Asaka, M.N. et al. Intratracheal trimerized nanobody cocktail medication suppresses value nonaccomplishment and prolongs endurance of SARS-CoV-2 infected mice. Commun Med 2, 152 (2022). https://doi.org/10.1038/s43856-022-00213-5
Received: 05 May 2022
Accepted: 08 November 2022
Published: 26 November 2022
DOI: https://doi.org/10.1038/s43856-022-00213-5




 English (US)
English (US)